Links
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
http://www.lulu.com/items/volume_70/10723000/10723072/1/print/10723072.pdf
http://www.jogg.info/52/files/Klyosov2.pdf http://www.scribd.com/doc/27979749/DNA-Genealogy-Mutation-Rates-And-Some-Historical-Evidence-Written-in-Y-Chromosome-Part-II-Walking-the-Map http://aklyosov.home.comcast.net/~aklyosov/ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Editorial Introduction
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
There goes a cliche that R1a1 haplotype is a Indo-European marker, associated exclusively with the Indo-Europeans. They have a patent on R1a1, and found anywhere, it is an undisputable evidence of Indo-European language. Or, rather, language and genetics. However, a closer examination allows to reach different conclusions. In Europe, the R1a figure may be approximately 60% of the European and of the 40% Asian origin, and who holds the patent and who is a knock-off is a matter of parochial patriotism. In Asia, the figure may be reversed, say 90% Asian origin, and 10% European. The highest R1 diversity is in Pakistan, maybe it was a refuge 18,000 ybp, or maybe it is a more recent crossroad where all versions of R1 deposited their genes. Generally speaking, no special training is needed to grasp the methods, logics, and results of the work.
The offered citation from the A. Klyosov article is discussing, among other things, the 3 manifestations of the R1a1, the first of which must be identical with the R1b, because R1b is a forking development within the R1 populace, and therefore the R1b1 Kurgans who invaded Europe from the west ca. 4000 BC must be (genetically) the same as the R1a1 who reached the Balkans ca. 10-8 mill. BC. In other words, from the demographic standpoint they are the same people marked by different genetic markers, distanced by 5 millennia and technology of horse husbandry, who admixed to different existing populations. And their language(s) must be genetically related, i.e. the Balkans' R1a1 spoke a Proto-language of the Iberian Kurgans R1b1, or a Proto-Proto-Bask/Celtic/Italic agglutinative vernacular BEFORE they entered Balkans, and Proto-non-IE, the language of the Old Europe, AFTER they started westward expansion under the pressure of R1b1 Kurganians ca. 4300 BC. The Iberian Kurgans R1b1 with Proto-Bask/Celtic/Italic agglutinative vernacular BEFORE they entered Northern Europe were already speaking the Proto-Proto-IE language BEFORE they expanded to the Eastern Europe as Corded Ware, and Proto-IE language AFTER they expanded to Urals, Central Asia, and eventually India. The linguistic change from R1a1 and R1b1 agglutinative to R1a1 flexive took place in the Northern Europe between 4th and 3rd mill. BC, and therefore the first “Indian” R1a1's from S.Siberia before the 5th mill. BC spoke the same Proto-Proto-Bask/Celtic/Italic agglutinative vernacular as the Balkan invaders ca. 10-8 mill. BC, close in time to the R1a1 Balkans and to the R1b1 Kurganians. The second “Indian” R1a1's from Central Asia spoke a different, the new flexive Proto-IE language.
In the author's terminology, Russians are Slavs, and Slavs come in 3 flavors, Eastern Slavs, Western Slavs, and Northern (or Uralic) Slavs. A more detailed definition can be found in http://www.springerlink.com/content/17w1713787p66m70/:
“... ethnic Russians, that is Eastern Slavs (R1a1 tribe), Western Slavs (I1 and I2 tribes), and Northern (or Uralic) Slavs (N1c tribe), which were found to live around 4600 years before present (R1a1), 3650 ybp (I1), 3000 and 10,500 ybp (I2, two principal DNA lineages), and 3525 ybp (N1c)....” In that context, Russians are descendents of R1a1 Siberians who in the genetical record first appear 20 mill. ybp in Siberia and next appear 12-10 mill. ybp in the Balkans. Their original language can first be detected by the languages of their Kurgan kins in the Iberia at about 4000 BC:
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Anatole A. Klyosov
DNA Genealogy, Mutation Rates, And Some Historical Evidence Written in Y-Chromosome, Part II: Walking the map | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
217
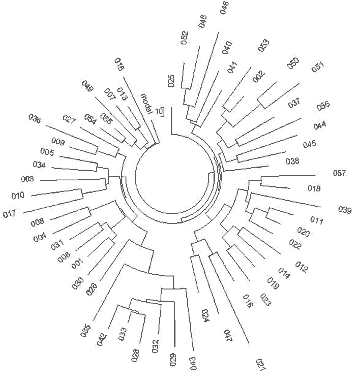
Summary218 The data presented below will show several major conclusions: (a) The male Basques living today have rather recent roots of less than four thousand years ago, contrary to the legend that proposes they lived some 30 thousand years ago. (b) There is no justification in the results of a “Ukrainian refuge” for the R1a1 ancient population allegedly 15,000 years ago; instead, evidence has been obtained that the oldest R1a1 lived circa 20,000 years before the present (ybp) in South Siberia. There are two sets of data and these provide ages of 21,000±3,000 ybp and 19,625±2,800 ybp, calculated by two different methods, and 11,650±1,550 years ago they appeared in the Balkans (Serbia, Kosovo, Bosnia, Macedonia). (c) Except the South Siberian and Balkans populations, present-day bearers of R1a1 across Western and Eastern Europe have common ancestors who lived between 3550 and 4750 years ago (the “youngest” in Scotland, Ireland and Sweden, the “oldest” in Russia (4750±500 ybp) and Germany (4,700±520 ybp), (d) There are two different groups of Indian R1a1 haplotypes; one shows a good match with the Russian Slavic R1a1 group, having a common ancestor several hundred years “younger” than the Russian R1a common ancestor (4,050±500 vs. 4,750±500 ybp). This supports the idea that a proto-Slavic migration to India as Aryans occurred (mentioned in classic ancient Indian literature) around 3600 ybp. The other Indian R1a population is significantly older, with a common ancestor living 7,125±950 ybp; they could have migrated from South Siberia to South India. (e) South India Chenchu R1a1 match the current Russian Slavic R1a1 haplotypes, and the Chenchu R1a common ancestor appeared some 3200±1900 ybp, apparently after the R1a1 migration from the North to India. Another Chenchu R1a1 lineage originated about 350±350 ybp, around the 17th century CE. (f) The so-called Cohen Modal Haplotype in Haplogroup J1 originated 9,000±1,400 years ago, if all related J1 haplotypes are considered. About 4,000±520 ybp it appeared in the proto-Jewish population, and 1,050±190 years ago (if to consider only CMH) or 1,400±260 years ago (if to consider only Jewish J1 population) split a “recent CMH” lineage. (g) another so-called CMH, of Haplogroup J2, appeared in the Jewish population 1,375±300 years ago, (h)The South African Lemba population of Haplogroup J has nothing to do with ancient Jewish patriarchs, since the haplogroup appears to have penetrated the Lemba population some 625±200 ybp, around the 14th century CE. (i) Native American Haplogroup Q1a3a contains at least six lineages, the oldest of which originated 16,000±3,300 years ago, in accord with archaeological data. The methods used in the present study are described in the companion article. Results and Discussion: Eurasian R1a1 Populations The “mapping” of the enormous territory from the Atlantic through Russia and India to the Pacific, and from Scandinavia to the Arabian Peninsula, reveals that Haplogroup R1a men are marked with practically the same ancestral haplotype, which is about 4,500 - 4,700 years “old.” through much of its geographic range. Exceptions in Europe are found only in the Balkans (Serbia, Kosovo, Macedonia, Bosnia), where the common ancestor is significantly more ancient, about 11,650±1,550 ybp, and in the Irish, Scottish and Swedish R1a1 populations, which have a significantly “younger” common ancestor, some thousands years “younger” compared with the Russian, the German, and the Poland R1a1 populations. These geographic patterns will be explored below in this section. The haplogroup name R1a1 is used here to mean Haplogroup R-M17, because that is the meaning in nearly all of the referenced articles. The entire map of base (ancestral) haplotypes and their mutations, as well as “ages” of common ancestors of R1a1 haplotypes in Europe, Asia, and the Middle East show that approximately six thousand years ago bearers of R1a1 haplogroup started to migrate from the Balkans in all directions, spreading their haplotypes. A recent excavation of 4,600 year-old R1a1 haplotypes (Haak et al., 2008) revealed their almost exact match to present day R1a1 haplotypes, as it is shown below. Significantly, the oldest R1a population appears to be in Southern Siberia. England and Ireland R1a1 haplotypes The 57 R1a 25-marker haplotype series of English origin (YSearch database) contains ten haplotypes that belong to a DYS388=10 series and was analyzed separately. The remaining 47 haplotypes contain 304 mutations compared to the base haplotype shown below, which corresponds to 4,125±475 years to a common ancestor in the 95% confidence interval. The respective haplotype tree is shown in Fig. 3. 225
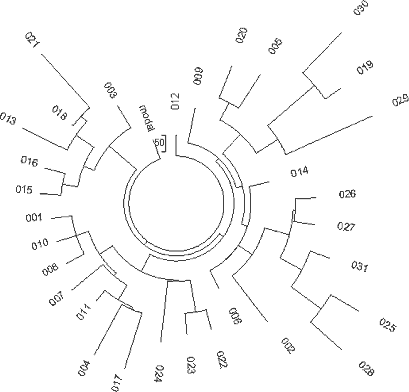
Thus, R1a1 haplotypes sampled on the British Isles point to English and Irish common ancestors who lived 4,125±475 and 3,850±460 years ago. The English base (ancestral) haplotype is as follows 13-25-15-10-11-14-12-12-10-13-11-30-15-9-10-11-11- 24-14 20-32-12-15-15-16 and the Irish one: 13-25-15- -11-14-12-12-10-13-11-30-15-9-10-11-11- -14 20-32-12-15-15-16 An apparent difference in two alleles between the British and Irish ancestral haplotypes is in fact fairly insignificant, since the respective average alleles are equal to 10.51 and 10.73, and 23.98 and 23.55, respectively. Hence, their ancestral haplotypes are practically the same, within approximately one mutational difference. A DYS388=10 Subfamily of North-Western European R1a1 Haplotypes About 20% of both English and Irish R1a haplotypes have a mutated allele in eighth position in the FTDNA format (DYS388=12à10), with a common ancestor of that population who lived 3,575±450 years ago (172 mutations in the 30 25-marker haplotypes with DYS388=10). 61 of these haplotypes were pooled from a number of European populations ( Fig. 5), and the tree splits into a relatively younger branch on the left, and the “older” branch on the lower right-hand side. 226
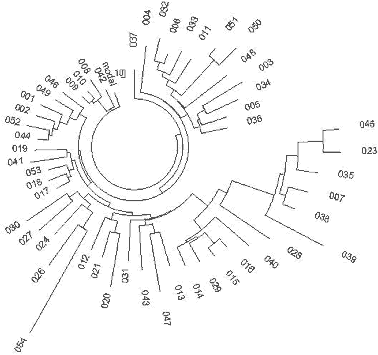
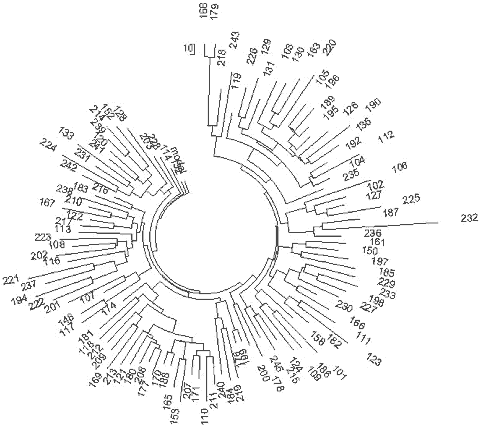
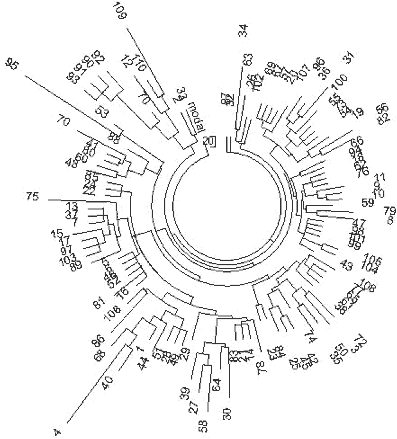
31 haplotypes on the left-hand side and on the top of the tree (Fig. 5) contain collectively 86 mutations from the base haplotype: 13-25-15-10-11-14-12-10-10-13-11-30-15-9-10-11-11- 25-14-19-32-12-14-14-17 which corresponds to 1625±240 years to the common ancestor. It is a rather recent common ancestor, who lived around the 4th century CE. The closeness of the branch to the trunk of the tree in also points to the rather recent origin of the lineage. The older 30-haplotype branch in the tree provides with the following base DYS388=10 haplotype: 13-25-16-10-11-14-12-10-10-13-11-30-15-9-10-11-11- 24-14-19-32-12-14-15-16 These 30 haplotypes contain 172 mutations, for which the linear method gives 3575±450 years to the common ancestor. These two DYS388=10 base haplotypes differ from each other by less than four mutations, which brings their common ancestor to about 3500 ybp. It is very likely that it is the same common ancestor as that of the right-hand branch in Fig. 5. The upper, “older” base haplotype differs by six mutations on average from the DYS388=12 base haplotypes from the same area (see the English and Irish R1a1 base haplotypes, above). This brings common ancestor to about 5,700±600 ybp. This common ancestor of both the DYS388=12 and DYS388=10 populations lived presumably in the Balkans (see below) since the Balkan population is the only European R1a1 population that is that old, for almost two thousand years before bearers of that mutation arrived to northern and western Europe some 4000 ybp (DYS388=12) and about 3600 ybp (DYS388=10). This mutation has continued to be passed down through the generations to the present time. 227 Scotland R1a1 Haplotypes A set of 29 R1a1 25-marker haplotypes from Scotland have the same ancestral haplotype as those in England and Ireland: 13-25-15-11-11-14-12-12-10-13-11-30-15-9-10-11-11- 24-14 20-32-12-15-15-16 This set of haplotypes contained 164 mutations, which gives 3550±450 years to the common ancestor. Germany R1a1 Haplotypes A 67-haplotype series with 25 markers from Germany revealed the following ancestral haplotype: 13-25-16-10-11-14-12-12-10-13-11-30-15-9-10-11-11- 24-14 20-32-12-15-15-16 There is an apparent mutation in the third allele from the left (in bold) compared with the Isles ancestral R1a1 haplotypes, however, the average value equals to 15.48 for England, 15.33 for Ireland, 15.26 for Scotland, and 15.84 for Germany, so there is only a small difference between these populations. The 67 haplotypes contain 488 mutations, or 0.291±0.013 mutations per marker on average. This value corresponds to 4,700±520 years from a common ancestor in the German territory. These results are supported by the very recent data found during excavation of remains from several males near Eulau, Germany. Tissue samples from one of them was derived for the SNP, SRY10831.2 (Haak et al., 2008), so it was from Haplogroup R1a1 and probably R1a1a as well (the SNPs defining R1a1a were not tested). The skeletons were dated to 4,600 ybp using strontium isotope analysis. The 4600 year old remains yielded some DNA and a few STRs were detected, yielding the following partial haplotype: 13(14)-25-16-11-11-14-X-Y-10-13-Z-30-15 These haplotypes very closely resemble the above R1a1 ancestral haplotype in Germany both in the structure and in the dating (4,700±520 and 4,600 ybp). Norway and Sweden R1a1 Haplotypes The ancestral R1a1 haplotypes for the Norwegian and Swedish groups are almost the same, and both are similar to the German 25-marker ancestral haplotype. Their 16-and 19-haplotype sets (after DYS388=10 haplotypes were removed, five and one, respectively) contain on average 0.218±0.023 and 0.242±0.023 mutations per marker, respectively, which gives a result of 3,375±490 and 3,825±520 years back to their common ancestors. This is likely the same time span within the error margin. Polish, Czech, and Slovak R1a1 Haplotypes The ancestral haplotypes for the Polish, Czech, and Slovak groups are very similar to each other, having only one insignificant difference in DYS439 (shown in bold). The average value is 10.43 on DYS439 in the Polish group, 10.63 in the Czech and Slovak combined groups. The base haplotype for the 44 Polish haplotypes is:. 13-25-16-10-11-14-12-12- -13-11-30-16-9-10-11-11- 23-14-20-32-12-15-15-16 and for the 27 Czech and Slovak haplotypes it is: 13-25-16-10-11-14-12-12- 13-11-30-16-9-10-11-11- 23-14-20-32-12-15-15-16 The difference in these haplotypes is really only 0.2 mutations - the alleles are just rounded in opposite directions. The 44 Polish haplotypes have 310 mutations among them, and there are 175 mutations in the 27 Czech-Slovak haplotypes, resulting in 0.282±0.016 and 0.259±0.020 mutations per marker on average, respectively. These values correspond to 4,550±520 and 4,125±430 years back to their common ancestors, respectively, which is very similar to the German TSCA. The Remaining European R1a1 Haplotypes Many European countries are represented by just a few 25-marker haplotypes in the databases. 36 R1a1 haplotypes were collected from the YSearch database where the ancestral origin of the paternal line was from Denmark, Netherlands, Switzerland, Iceland, Belgium, France, Italy, Lithuania, Romania, Albania, Montenegro, Slovenia, Croatia, Spain, Greece, Bulgaria and Moldavia. The respective haplotype tree is shown in Fig. 6. The tree does not show any noticeable anomalies and points at just one common ancestor for all 36 individuals, who had the following base haplotype: 13-25-16-10-11-14-12-12-10-13-11-30-15-9-10-11-11- 24-14-20-32-12-15-15-16 The ancestral haplotype match is exactly the same as those in Germany, Russia (see below), and has quite insignificant deviations from all other ancestral haplotypes considered above, within fractions of mutational differences. All the 36 individuals have 248 mutations in their 25-marker haplotypes, which corresponds to 4,425±520 years to the common ancestor. This is quite a common value for European R1a1 population. Russia and Ukraine R1a1 Haplotypes The haplotype tree containing 110 of 25-marker haplotypes collected over 10 time zones from the Western Ukraine to the Pacific Ocean and from the northern tundra to Central Asia (Tadzhikistan and Kyrgyzstan) is shown in Fig. 7. 228
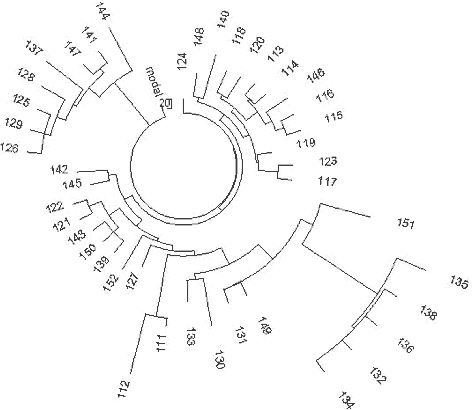
13-25-16-11-11-14-12-12-10-13-11-30-15-9-10-11-11- 24-14-20-32-12-15-15-16 It is almost exactly the same ancestral haplotype as that in Germany. The average value on DYS391 is 10.53 in the Russian haplotypes, and was rounded to 11, and the base haplotype has only one insignificant deviation from the ancestral haplotype in England, which has the third allele (DYS19) 15.48, which in Russia/Ukraine it is 15.83 (calculated from one DYS19=14, 32 DYS19=15, 62 DYS19=16, and 15 DYS19=17). There are similar insignificant deviations with the Polish and Czechoslovakian base haplotypes, at DYS458 and DYS447, respectively, within 0.2-0.5 mutations. This indicates a small mutational difference between their common ancestors. The 110 Russian/Ukranian haplotypes contain 804 mutations from the base haplotype, or 0.292±0.010 mutations per marker, resulting in 4,750±500 years from a common ancestor. The degree of asymmetry of this series of haplotypes is exactly 0.50, and does not affect the calculations. 229 R1a1 haplotypes of individuals who considered themselves of Ukrainian and Russian origin, present a practically random geographic sample. For example, the haplotype tree contains two local Central Asian haplotypes (a Tadzhik and a Kyrgyz, haplotypes 133 and 127, respectively), as well as a local of a Caucasian Mountains Karachaev tribe (haplotype 166), though the male ancestry of the last one is unknown beyond his present tribal affiliation. They did not show any unusual deviations from other R1a1 haplotypes. Apparently, they are derived from the same common ancestor as are all other individuals of the R1a1 set. The literature frequently refers to a statement that R1a1- M17 originated from a “refuge” in the present Ukraine about 15,000 years ago, following the Last Glacial Maximum. This statement was never substantiated by any actual data related to haplotypes and haplogroups. It is just repeated over and over through a relay of references to references. The oldest reference is apparently that of Semino et al. (2000) which states that “this scenario is supported by the finding that the maximum variation for microsatellites linked to Eu19 [R1a1] is found in Ukraine” (Santachiara-Benerecetti, unpublished data). Now we know that this statement is incorrect. No calculations were provided in (Semino et al, 2000) or elsewhere which would explain the dating of 15,000 years. Then, a paper by Wells et al. (2001) states “M17, a descendant of M173, is apparently much younger, with an inferred age of ~ 15,000 years.” No actual data or calculations are provided. The subsequent sentence in the paper says, “It must be noted that these age estimates are dependent on many, possibly invalid, assumptions about mutational processes and population structure.” This sentence has turned out to be valid in the sense that the estimate was inaccurate and overestimated by about 300% from the results obtained here. However, see the results below for the Chinese and southern Siberian haplotypes, below. 230 A more detailed consideration of R1a1 haplotypes in the Eastern European Plain and across Europe and Eurasia in general has shown that R1a1 haplogroup appeared in Europe between 12 and 10 thousand years before present, right after the Last Glacial Maximum, and after about 6,000 ybp had populated Europe, though, probably, with low density. After 4,500 ybp R1a1 practically disappeared from Europe, incidentally, along with I1. Maybe more incidentally, it corresponded with the time period of populating of Europe with R1b1b2. Only those R1a1 who migrated to the Eastern European Plain from Europe around 6,000-5,000 ybp, stayed. They had expanded to the East, established on their way a number of archaeological cultures, including the Andronovo culture, which has embraced Northern Kazakhstan, Central Asia and South Ural and Western Siberia, and about 3600 ybp they migrated to India and Iran as the Aryans. Those who left behind, on the Eastern European Plain, re-populated Europe between 3200 and 2500 years bp, and stayed mainly in the Eastern Europe (present-day Poland, Germany, Czech, Slovak, etc. regions). Among them were carriers of the newly discovered R1a1-M458 subclade (Underhill et al, 2009).
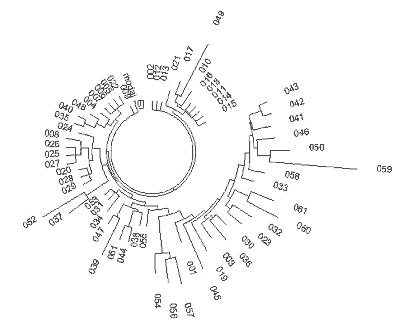
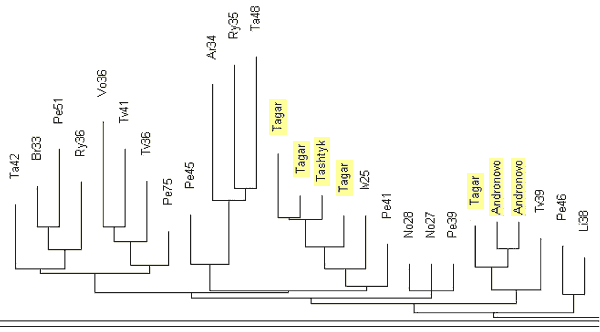
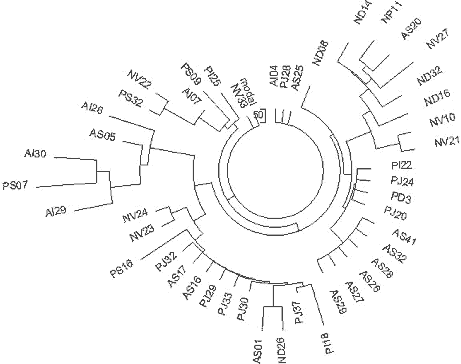
India R1a1 Haplotypes The YSearch database contains 22 of the 25-marker R1a1 haplotypes from India, including a few haplotypes from Pakistan and Sri Lanka. Their ancestral haplotype follows: 13-25-16-10-11-14-12-12-10-13-11-30-16-9-10-11-11- 24-14-20-32-12-15-15-16 The only one apparent deviation in DYS458 (shown in bold) is related to an average alleles equal to 16.05 in Indian haplotypes, and 15.28 in Russian ones. The India (Regional) Y-project at FTDNA (Rutledge, 2009) contains 15 haplotypes with 25 markers, eight of which are not listed in the YSearch database. Combined with others from Ysearch, a set of 30 haplotypes is available. Their ancestral haplotype is exactly the same as shown above. Fig. 8 shows the respective haplotypes tree. All 30 Indian R1a1 haplotypes contain 191 mutations, that is 0.255±0.018 mutations per marker. It is statistically lower (with 95% confidence interval) than 0.292±0.014 mutations per marker in the Russian haplotypes and corresponds to 4,050±500 years from a common ancestor of the Indian haplotypes, compared to 4,750±500 years for the ancient “Russian” TSCA. Archaeological studies have been conducted since the 1990’s in the South Ural’s Arkaim settlement and have revealed that the settlement was abandoned 3,600 years ago. The population apparently moved to northern India. That population belonged to Andronovo archaeological culture. Excavations of some sites of Andronovo culture, the oldest dating between 3,800 and 3,400 ybp, showed that nine inhabitants out of ten shared the R1a1 haplogroup and haplotypes (Bouakaze et al., 2007; Keyser et al, 2009). The base haplotype is as follows: 13-25(24)-16(17)-11-11-14-X-Y-10-13(14)-11-31(32) In this example, alleles that have not been assessed are replaced with letters. One can see that the ancient R1a1 haplotype closely resembles the Russian (as well as the other R1a1) ancestral haplotypes. 232 In a recent paper (Keyser et al, 2009) the authors have extended the earlier analysis and determined 17-marker haplotypes for 10 Siberian individuals assigned to Andronovo, Tagar, and Tashtyk archaeological cultures (See Keyser et al, 2009 Sibirian Kurgan Y- and mt-DNA
As one can see, the ancient R1a1 haplotypes excavated in Siberia, are comfortably located on the tree next to the haplotypes from the Russian cities and regions named in the legend to Fig. 10. The above data provide rather strong evidence that the R1a1 tribe migrated from Europe to the East between 5,000 and 3,600 ybp. The pattern of this migration is exhibited as follows: 1) the descendants who live today share a common ancestor of 4,725±520 ybp, 2) the Andronovo (and the others) archaeological complex of cultures in North Kazakhstan and South and Western Siberia dates 4,300 to 3,500 ybp, and it revealed several R1a1 excavated haplogroups, 3) they reached the South Ural region some 4,000 ybp, which is where they built Arkaim, Sintashta (contemporary names), and the so called “a country of towns” in the South Ural region around 3,800 ybp, 4) by 3,600 ybp they abandoned the area and moved to India under the name of Aryans. The Indian R1a1 common ancestor of 4,050±500 ybp chronologically corresponds to these events. Currently, some 16% of the Indian population, that is about 100 millions males, and the majority of the upper castes, are members of Haplogroup R1a1 (Sengupta et al, 2006; Sharma et al, 2009). Since we have mentioned Haplogroup R1a1 in India and in the archaeological cultures north of it, it is worthwhile to consider the question of where and when R1a1 haplotypes appeared in India. On the one hand, it is rather obvious from the above, that R1a1 haplotypes were brought to India around 3500 ybp from what is now Russia. The R1a1 bearers, known later as the Aryans, brought to India not only their haplotypes and the haplogroup, but also their language, thereby closing the loop, or building the linguistic and cultural bridge between India (and Iran) and Europe, and possibly creating the Indo-European family of languages. On the other hand, there is evidence that some Indian R1a1 haplotypes show a high variance, exceeding that in Europe (Kivisild et al, 2003; Sengupta et al, 2006; Sharma et al, 2009; Thanseem et al, 2009; Fornarino et al, 2009), thereby antedating the 4000-year-old migration from the north.However, there is a way to resolve the conundrum. 233 It seems that there are two quite distinct sources of Haplogroup R1a1 in India. One, indeed, was probably brought from the north by the Aryans. However, the most ancient source of R1a1 haplotypes appears to be provided by people who now live in China. In an article by Bittles et al (2007) entitled “Physical anthropology and ethnicity in Asia: the transition from anthropometry to genome-based studies” a list of frequencies of Haplogroup R1a1 is given for a number of Chinese populations, however, haplotypes were not provided. The corresponding author, Professor Alan H. Bittles, kindly sent me the following list of 31 five-marker haplotypes (presented here in the format DYS19, 388, 389-1, 389-2, 393), in Table 3. The haplotype tree is shown in Fig. 11.
13-X-14-X-X-X-X-12-X-13-X-30 which gives 0.639±0.085 mutations per marker, or 21,000±3,000 years to a common ancestor. Such a large timespan to a common ancestor results from a low mutation rate constant, which was calculated from the Chandler’s data for individual markers, as 0.00677 mutation/haplotype/generation, or 0.00135 mutation/marker/generation (see Table 1 in Part I of the companion article). Since haplotypes descended from such a ancient common ancestor have many mutations, which makes their base (ancestral) haplotypes rather uncertain, the ASD permutational method was employed for the Chinese set of haplotypes (Part I). The ASD permutational method does not need a base haplotype and it does not require a correction for back mutations. For the given series of 31 five-marker haplotypes the sum of squared differences between each allele in each marker equals to 10,184. It should be divided by the square of a number of haplotypes in the series (961), by a number of markers in the haplotype (5) and by 2, since the squared differences between alleles in each marker were taken both ways. It gives an average number of mutations per marker of 1.060. After division of this value by the mutation rate for the five-marker haplotypes (0.00135 mut/marker/generation for 25 years per generation), we obtain 19,625±2,800 years to a common ancestor. It is within the margin of error with that calculated by the linear method, as shown above. It is likely that Haplogroup R1a1 had appeared in South Siberia around 20 thousand years ago, and its bearers split. One migration group headed West, and had arrived to the Balkans around 12 thousand years ago (see below). Another group had appeared in China some 21 thousand years ago. Apparently, bearers of R1a1 haplotypes made their way from China to South India between five and seven thousand years ago, and those haplotypes were quite different compared to the Aryan ones, or the “Indo-European” haplotypes. This is seen from the following data.
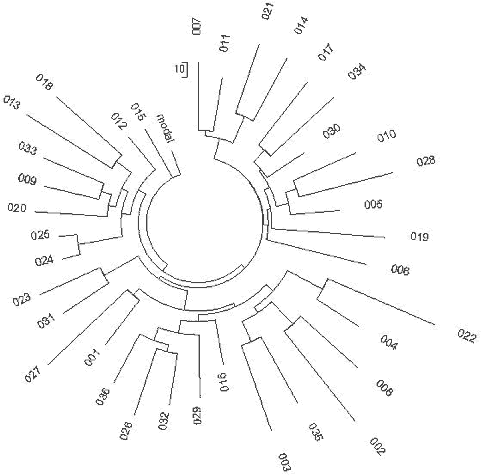
13-25-17-9-X-X-X-X-X-14-X-32 It differs from the “Indo-European” Indian haplotype, 13-25-16-10-11-14-12-12-10-13-11-30 by four mutations on six markers, which corresponds to 11,850 years between their common ancestors, and places their common ancestor at approximately (11850+4050+7125)/2 = 11,500 ybp. 236 110 of 10-marker R1a1 haplotypes of various Indian populations, both tribal and Dravidian and Indo-European castes, listed in (Sengupta et al, 2006) shown in a haplotype tree in Figure 13, contain 344 mutations, that is 0.313±0.019 mutations per marker. It gives 5,275±600 years to a common ancestor. The base (ancestral) haplotype of those populations in the FTDNA format is as follows: 13-25-15-10-X-X-X-12-10-13-11-30 It differs from the “Indo-European” Indian haplotype 13-25-16-10-11-14-12-12-10-13-11-30 by just 0.5 mutations on nine markers (DYS19 has, in fact, a mean value of 15.5 in the Indian haplotypes), which makes them practically identical. However, in this haplotype series two different populations, the “Indo-European” one and the “South-Indian” one were mixed, therefore an “intermediate”, apparently phantom “common ancestor” was artificially created with an intermediate TSCA, between 4,050±500 and 7,125±950 (see above). For a comparison, let us consider Pakistani R1a1 haplotypes listed in the article by Sengupta (2003) and shown in Fig. 14. The 42 haplotypes contain 166 mutations, which gives 0.395±0.037 mutations per marker, and 7,025±890 years to a common ancestor. This value fits within the margin of error to the “South-Indian” TSCA of 7,125±950 ybp. The base haplotype was as follows: 13-25-17-11-X-X-X-12-10-13-11-30 It differs from the “Indo-European” Indian haplotype by two mutations in the nine markers.
237 The above suggests that there are two different subsets of Indian R1a1 haplotypes. One was brought by European bearers known as the Aryans, seemingly on their way through Central Asia, in the 2nd millennium BCE, another, much more ancient, made its way apparently through China, and arrived in India earlier than seven thousand years ago. R1a1 Haplotypes of the Arabian Peninsula Sixteen R1a1 10-marker haplotypes from Qatar and United Arab Emirates have been recently published (Cadenas et al., 2008). They split into two branches, with base haplotypes 13-25-15-11-11-14-X-Y-10-13-11-30 13-25-16-11-11-14-X-Y-10-13-11-31 which are only slightly different, and on DYS19 the mean values are almost the same, just rounded up or down. The first haplotype is the base one for seven haplotypes with 13 mutations in them, on average 0.186±0.052 mutations per marker, which gives 2,300±680 years to a common ancestor. The second haplotype is the base one for nine haplotypes with 26 mutations, an average 0.289±0.057 mutations per marker or 3,750±825 years to a common ancestor. Since a common ancestor of R1a1 haplotypes in Armenia and Anatolia lived 4,500±1,040 and 3,700±550 ybp, respectively (Klyosov, 2008b), it does not conflict with 3,750±825 ybp in the Arabian peninsula. The Balkan Ancient Branch: the Oldest Trace of R1a Haplotype in Europe? A series of 67 haplotypes of Haplogroup R1a1 from the Balkans was published (Barac et al., 2003a, 2003b; Pericic et al., 2005). They were presented in a 9-marker format only. The respective haplotype tree is shown in Fig. 15. 238 One can see a remarkable branch on the left-hand side of the tree which stands out as an “extended and fluffy” one. These are typically features of a very old branch compared with others on the same tree. Also, a common feature of ancient haplotype trees is that they are typically “heterogeneous” ones and consist of a number of branches. The tree in Fig. 15 includes a rather small branch of twelve haplotypes on top of the tree, which contains only 14 mutations. This results in 0.130±0.035 mutations per marker, or 1,850±530 years to a common ancestor. Its base haplotype 13-25-16-10-11-14-X-Y-Z-13-11-30 is practically the same as that in Russia and Germany.
13-25-16-11-11-14-X-Y-Z-13-11-30 is again typical for Eastern European R1a1 base haplotypes, in which the fourth marker (DYS391) often fluctuates between 10 and 11. Of 110 Russian-Ukrainian haplotypes diagramed in Fig. 7, 51 haplotypes have “10”, and 57 have “11” in that locus (one has “9” and one has “12”). In 67 German haplotypes, discussed above, 43 haplotypes have “10”, 23 have “11” and one has “12.” Hence, the Balkan haplotypes from this branch are more close to the Russian than to the German haplotypes.
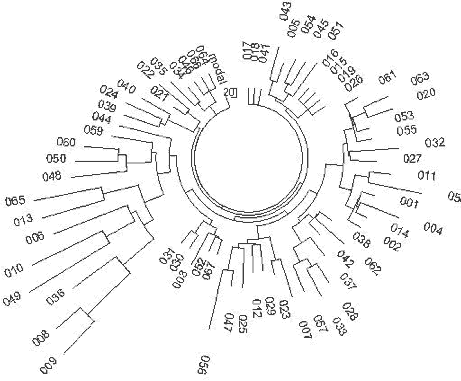
13 24 16 12 14 15 13 11 31 12 24 16 10 12 15 13 13 29 12 24 15 11 12 15 13 13 29 14 24 16 11 11 15 15 11 32 13 23 14 10 13 17 13 11 31 13 24 14 11 11 11 13 13 29 13 25 15 9 11 14 13 11 31 13 25 15 11 11 15 12 11 29 12 22 15 10 15 17 14 11 30 14 25 15 10 11 15 13 11 29 13 25 15 10 12 14 13 11 29 13 26 15 10 11 15 13 11 29 13 23 15 10 13 14 12 11 28 The set does not contain a haplotype which can be defined as a base. This is because common ancestor lived too long ago, and all haplotypes of his descendants living today are extensively mutated. In order to determine when that common ancestor lived, we have employed three different approaches, described in the preceding paper (Part 1), namely the “linear” method with the correction for reverse mutations, the ASD method based on a deduced base (ancestral) haplotype, and the permutational ASD method (no base haplotype considered). The linear method gave the following deduced base haplotype, an alleged one for a common ancestor of those 13 individuals from Serbia, Kosovo, Bosnia and Macedonia: 13-24-15-10-12-15-X-Y-Z-13-11-29 The bold notations identify deviations from typical ancestral (base) East-European haplotypes. The third allele (DYS19) is identical to the Atlantic and Scandinavian R1a1 base haplotypes. All 13 haplotypes contain 70 mutations from this base haplotype, which gives 0.598±0.071 mutations on average per marker, and results in 11,425±1,780 years from a common ancestor. The “quadratic method” (ASD) gives the following “base haplotype” (the unknown alleles are eliminated here, and the last allele is presented as the DYS389-2 notation): 12.92 – 24.15 – 15.08 – 10.38 – 12.08 – 14.77 – 13.08 – 11.46 – 16.62 A sum of square deviations from the above haplotype results in 103 mutations total, including reverse mutations “hidden” in the linear method. Seventy “observed” mutations in the linear method amount to only 68% of the “actual” mutations including reverse mutations. Since all 13 haplotypes contain 117 markers, the average number of mutations per marker is 0.880±0.081, which corresponds to 0.880/0.00189 = 466±62 generations or 11,650±1,550 years to a common ancestor. 0.00189 is the mutation rate (in mutations per marker per generation) for the given 9-marker haplotypes (companion paper, Part I). A calculation of 11,650±1,550 years to a common ancestor is practically the same as 11,425±1,780 years, obtained with linear method and corrected for reverse mutations. The all-permutation “quadratic” method (Adamov & Klyosov, 2008) gives 2,680 as a sum of all square differences in all permutations between alleles. When divided by N2 (N = number of haplotypes, that is 13), by 9 (number of markers in haplotype), and by 2 (since deviation were both “up” and “down”), we obtain an average number of mutations per marker equal to 0.881. It is near exactly equal to 0.880 obtained by the quadratic method above. Naturally, it gives again 0.881/0.00189 = 466±62 generations or 11,650±1,550 years to a common ancestor of the R1a1 group in the Balkans. 239 These results suggest that the first bearers of the R1a1 haplogroup in Europe lived in the Balkans (Serbia, Kosovo, Bosnia, Macedonia) between 10 and 13 thousand ybp. It was shown below that Haplogroup R1a1 has appeared in Asia, apparently in China or rather South Siberia, around 20,000 ybp. It appears that some of its bearers migrated to the Balkans in the following 7-10 thousand years. It was found (Klyosov, 2008a) that Haplogroup R1b appeared about 16,000 ybp, apparently in Asia, and it also migrated to Europe in the following 11-13 thousand years. It is of a certain interest that a common ancestor of the ethnic Russians of R1b is dated 6775±830 ybp (Klyosov, to be published), which is about two to three thousand years earlier than most of the European R1b common ancestors. It is plausible that the Russian R1b have descended from the Kurgan archaeological culture.
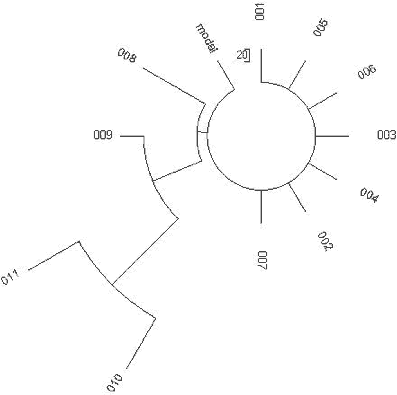
The Chenchu R1a1 Haplotypes Kivisild et al. (2003) reported that eleven out of 41 individuals tested in the Chenchu, an australoid tribal group from southern India, are in Haplogroup R1a1 (or 27% of the total). It is tempting to associate this with the Aryan influx into India, which occurred some 3,600-3,500 ybp. However, questionable calculations of time spans to a common ancestor of R1a1 in India, and particularly in the Chenchu (Kivisild et al., 2003; Sengupta et al., 2006; Thanseem et al, 2006; Sahoo et al, 2006; Sharma et al, 2009; Fornarino et al, 2009) using methods of population genetics rather than those of DNA genealogy have precluded an objective and balanced discussion of the events and their consequences. The eleven R1a1 haplotypes of the Chenchus (Kivisild et al., 2003) do not provide good statistics; however, they can allow a reasonable estimate of a time span to a common ancestor for these 11 individuals. Logically, if these haplotypes are more or less identical, with just a few mutations in them, a common ancestor would likely have lived within a thousand or two thousands of ybp. Conversely, if these haplotypes are all mutated, and there is no base (ancestral) haplotype among them, a common ancestor lived thousands ybp. Even two base (identical) haplotypes among 11 would tentatively give ln(11/2)/0.0088 = 194 generations, which, corrected to back mutations, would result in 240 generations, or 6,000 years to a common ancestor, with a large margin of error. If eleven of the six-marker haplotypes are all mutated, it would mean that a common ancestor lived apparently earlier than 6 thousand ybp. Hence, even with such a small set of haplotypes one can obtain useful and meaningful information. The eleven Chenchu haplotypes have seven identical (base) six-marker haplotypes (in the format of DYS 19-388-290-391-392-393, commonly employed in earlier scientific publications): 16-12-24-11-11-13 They are practically the same as those common East European ancestral haplotypes considered above, if presented in the same six-marker format: 16-12-25-11(10)-11-13 Actually, the author of this study, himself an R1a1 (Slav R1a1) has the “Chenchu” base six-marker haplotype. These identical haplotypes are represented by a “comb” in Fig 16. If all seven identical haplotypes are derived from the same common ancestor as the other four mutated haplotypes, the common ancestor would have lived on average of only 51 generations bp, or less than 1300 years ago [ln(11/7)/0.0088 = 51], with a certain margin of error (see estimates below). In fact, the Chenchu R1a1 haplotypes represent two lineages, one 3,200±1,900 years old and the other only 350±350 years old, starting from around the 17th century CE. The tree in Fig 16 shows these two lineages. 240
15-12-25-10-11-13 The recent branch results in ln(8/7)/0.0088 = 15 generations (by the logarithmic method), and 1/8/0.0088 = 14 generations (the linear method) from the residual seven base haplotypes and a number of mutations (just one), respectively. It shows a good fit between the two estimates. This confirms that a single common ancestor for eight individuals of the eleven lived only about 350±350 ybp, around the 17th century. The old branch of haplotypes points at a common ancestor who lived 3/3/0.0088 = 114±67 generations BP, or 3,200±1,900 ybp with a correction for back mutations. 241 Considering that the Aryan (R1a1) migration to northern India took place about 3,600-3,500 ybp, it is quite plausible to attribute the appearance of R1a1 in the Chenchu by 3,200±1,900 ybp to the Aryans. The origin of the influx of Chenchu R1a1 haplotypes around the 17th century is likely found in this passage excerpted from (Kivisild et al., 2003): “Chenchus were first described as shy hunter-gatherers by the Mohammedan army in 1694.” Native American haplotypes of Haplogroup Q1a3a http://s155239215.onlinehome.us/turkic/60_Genetics/Klyosov/Klyosov2009R1aDNAEn.htm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Темы
C
Cеквенирование
E
E1b1b
G
I
I1
I2
J
J1
J2
N
N1c
Q
R1a
R1b
Y-ДНК
Австролоиды
Альпийский тип
Америнды
Англия
Антропологическая реконструкция
Антропоэстетика
Арабы
Арменоиды
Армия Руси
Археология
Аудио
Аутосомы
Африканцы
Бактерии
Балканы
Венгрия
Вера
Видео
Вирусы
Вьетнам
Гаплогруппы
Генетика человека
Генетические классификации
Геногеография
Германцы
Гормоны
Графики
Греция
Группы крови
ДНК
Деградация
Демография в России
Дерматоглифика
Динарская раса
Дравиды
Древние цивилизации
Европа
Европейская антропология
Европейский генофонд
ЖЗЛ
Живопись
Животные
Звёзды кино
Здоровье
Знаменитости
Зодчество
Иберия
Индия
Индоарийцы
Интеръер
Иран
Ирландия
Испания
Исскуство
История
Италия
Кавказ
Канада
Карты
Кельты
Китай
Корея
Криминал
Культура Руси
Латинская Америка
Летописание
Лингвистика
Миграция
Мимикрия
Мифология
Модели
Монголоидная раса
Монголы
Мт-ДНК
Музыка для души
Мутация
Народные обычаи и традиции
Народонаселение
Народы России
Наши Города
Негроидная раса
Немцы
Нордиды
Одежда на Руси
Ориентальная раса
Основы Антропологии
Основы ДНК-генеалогии и популяционной генетики
Остбалты
Переднеазиатская раса
Пигментация
Политика
Польша
Понтиды
Прибалтика
Природа
Происхождение человека
Психология
РАСОЛОГИЯ
РНК
Разное
Русская Антропология
Русская антропоэстетика
Русская генетика
Русские поэты и писатели
Русский генофонд
Русь
США
Семиты
Скандинавы
Скифы и Сарматы
Славяне
Славянская генетика
Среднеазиаты
Средниземноморская раса
Схемы
Тохары
Тураниды
Туризм
Тюрки
Тюрская антропогенетика
Укрология
Уралоидный тип
Филиппины
Фильм
Финляндия
Фото
Франция
Храмы
Хромосомы
Художники России
Цыгане
Чехия
Чухонцы
Шотландия
Эстетика
Этнография
Этнопсихология
Юмор
Япония
генетика
интеллект
научные открытия
неандерталeц